Host-Microbe Interactions
Bacterial infections constitute a major cause of morbidity and mortality worldwide and pose a significant threat to human health as the spread of antibiotic resistance increases. We are interested in the molecular crosstalk that takes place between pathogenic bacteria and host cells during different stages of infection. What are the molecular determinants underlying bacterial adhesion to- and invasion of host tissue, and how do infecting pathogens adapt to the often hostile environment inside a host organism? In order to address these questions we take a multidisciplinary approach involving bacterial genetics, biochemistry, microscopy, flow cytometry and proteomics for examination of simple infection model systems.
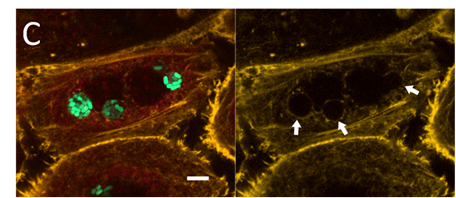
Figure 1: Intracellular UPEC colonies in bladder cells. GFP-expressing bacteria multiply inside vacuoles in the host cell cytoplasm.
Head of Research: Associate Professor Jakob Møller-Jensen
Research Group: Jakob Møller-Jensen Lab
Academic staff Mark Lyng
Postdoc Sergi Torres Puig
Guest researcher Anders Boysen
Guest researcher Mette Thorsing
Current Research Projects:
Genome-wide mapping of host-induced bacterial genes
In order to identify bacterial genes that are specifically turned on inside the host during infection, we employ a genetic method termed Differential Fluorescence Induction (DFI). This approach allows for selective enrichment and identification of in vivo-induced or repressed bacterial genes. In several cases host-induced genes have been shown to be required for the ability of pathogenic bacteria to successfully establish an infection. DFI is based on bacterial genetic libraries that are used for infection in tissue culture or live animals. Host cells containing GFP-fluorescing bacteria can be isolated at the relevant stage of the infection using fluorescence-activated cell sorting (FACS). In order to obtain a genome-wide rather than clonal readout we attempt to couple DFI with next generation sequencing analysis, a strategy we term DFI-Seq.
Important aspects of bacterial virulence, including stress tolerance and virulence factor production, are controlled at the posttranscriptional level by small non-coding RNAs (sRNAs). The function and stability of many sRNAs depends on a small abundant RNA-binding protein, Hfq. The Hfq protein acts as a global posttranscriptional regulator of gene expression by binding to sRNAs and facilitating their interaction with cognate mRNA molecules. Using Hfq immunoprecipitation and RNA deep sequencing we seek to determine the regulatory RNA molecules which come into play at specific stages of bacterial infection.
The role of protein glycosylation in bacterial pathogenesis
Glycosylated proteins have been identified in several important human pathogens and are known to play essential roles in bacterial toxicity, biofilm formation, adhesion and invasion of host tissue – all of which are hallmarks of bacterial pathogenesis. In collaboration with the Martin Larsen group we have developed a novel method, termed BEMAP, which allows for selective enrichment and subsequent identification of glycoproteins. Using BEMAP we have obtained preliminary data demonstrating that protein glycosylation is more pronounced pathogenic Escherichia colicompared to commensal strains. We hypothesize that these sugar modifications play important roles in the establishment of bacterial infections in general. BEMAP relies on generic chemical principles and can be employed to identify glycosylated proteins from any organism. At an overall perspective, this proposal holds potential to advance our knowledge of protein glycosylation and the importance of these modifications in mediating host-pathogen interactions during a wide range of bacterial infections. We intend to map the entire glycoprotein complement (glycome) of clinically relevant bacterial species, including enterotoxigenic Escherichia coli (ETEC), and further determine the importance of protein glycosylation for pathogenesis. In addition we seek to identify host cell receptors engaged in interaction with bacterial glycoproteins.
Study of Escherichia coli uropathogenesis using an artificial bladder infection model
Urinary tract infection (UTI) caused by uropathogenic Escherichia coli (UPEC) is among the most frequently occurring bacterial infectious diseases in industrialized countries. In Denmark 200.000-300.000 people acquire a treatment-requiring UTI annually. UPEC infections are characterized by a high prevalence of recurrence and research has shown that a likely reason for this is the ability of UPEC to invade and replicate inside bladder cells, thereby shielding itself against immune responses and antibiotic therapy. Extensive research based on mouse models has revealed a complex UPEC pathogenesis pathway involving urothelium cell invasion, intracellular replication into intracellular bacterial communities (IBCs) and spreading or “fluxing” to neighboring cells. We have established a flow chamber based dynamic in vitro cystitis model which enables induction of human uropathogenesis under in vivo-like flow conditions. This system recapitulates major characteristics of UPEC uropathogenesis including bladder cell invasion, intracellular bacterial growth and fluxing of bacterial filaments. The bladder infection model system opens up possibilities for detailed analysis of bacterial gene expression during successive stages of UPEC infection.
Selected publications
Escherichia coli uropathogenesis in vitro: Invasion, cellular escape, and secondary infection analyzed in a human bladder cell infection model.
Andersen, T. E.; Khandige, S.; Madelung, M.; Brewer, J.; Kolmos, H. J.; Møller-Jensen, J., Infect. Immun., 2012, 80(5), 1858-67
c-terminally truncated derivatives of Escherichia coli Hfq are proficient in riboregulation.
Olsen, A. S.; Møller-Jensen, J.; Brennan, R. G.; Valentin-Hansen, P., J. Mol. Biol., 2010, 404(2), 173-82
Microbiology. Dynamic instability of a bacterial engine.
Møller-Jensen, J.; Gerdes, K.; Science, 2004, 306(5698), 987-9
A full list of publications by associate professor Jakob Møller-Jensen can be seen here.
